Our Research
We are interested in how the molecules on immune cells instruct specific responses. By learning how these molecules work, we can develop ways to manipulate them. This is critical when our natural immune responses are not sufficient.
Receptor Organisation
Molecules on the surface of immune cells are called ‘receptors’. They act as a conduit of signals from the outside enabling cells to communicate.
Our lab is interested in how the spatial organisation of immune receptors controls how they work. When immune cells interact, these molecules arrange into intricate patterns at the interaction sites. Evidence suggests this organisation is vital for their function. Understanding this process can lead to a new understanding of what happens when cells fail to communicate properly. We can use this knowledge to design new therapies that manipulate protein organisation.

Super-resolution TIRF microscopy image showing the immune receptors TIGIT (magenta) and TCR (green) in action at the immune synapse. The yellow bar shows a distance of 1 μm (Worboys et al, 2023).
Receptor signalling
Receptors transduce signals from the outside into the cell. Although this is known to be important, it is incompletely understood. We are interested in how receptors on immune cells transduce information with the ultimate aim being to modulate this.
We are interested in how an immune cell called a T cell is regulated. T cells are tightly controlled by the T cell receptor. It recognises molecular patterns from foreign pathogens or cancers. Other receptors on T cells can fine tune T cell activation. Some of these receptors, immune checkpoints, prevent and regulate cellular activation. Blocking these receptors can fire up an immune response and is used to treat cancer.
We are particularly interested in how the immune checkpoint TIGIT regulates T cell activation. This understanding will help us determine in which contexts TIGIT blockade will be clinically effective.
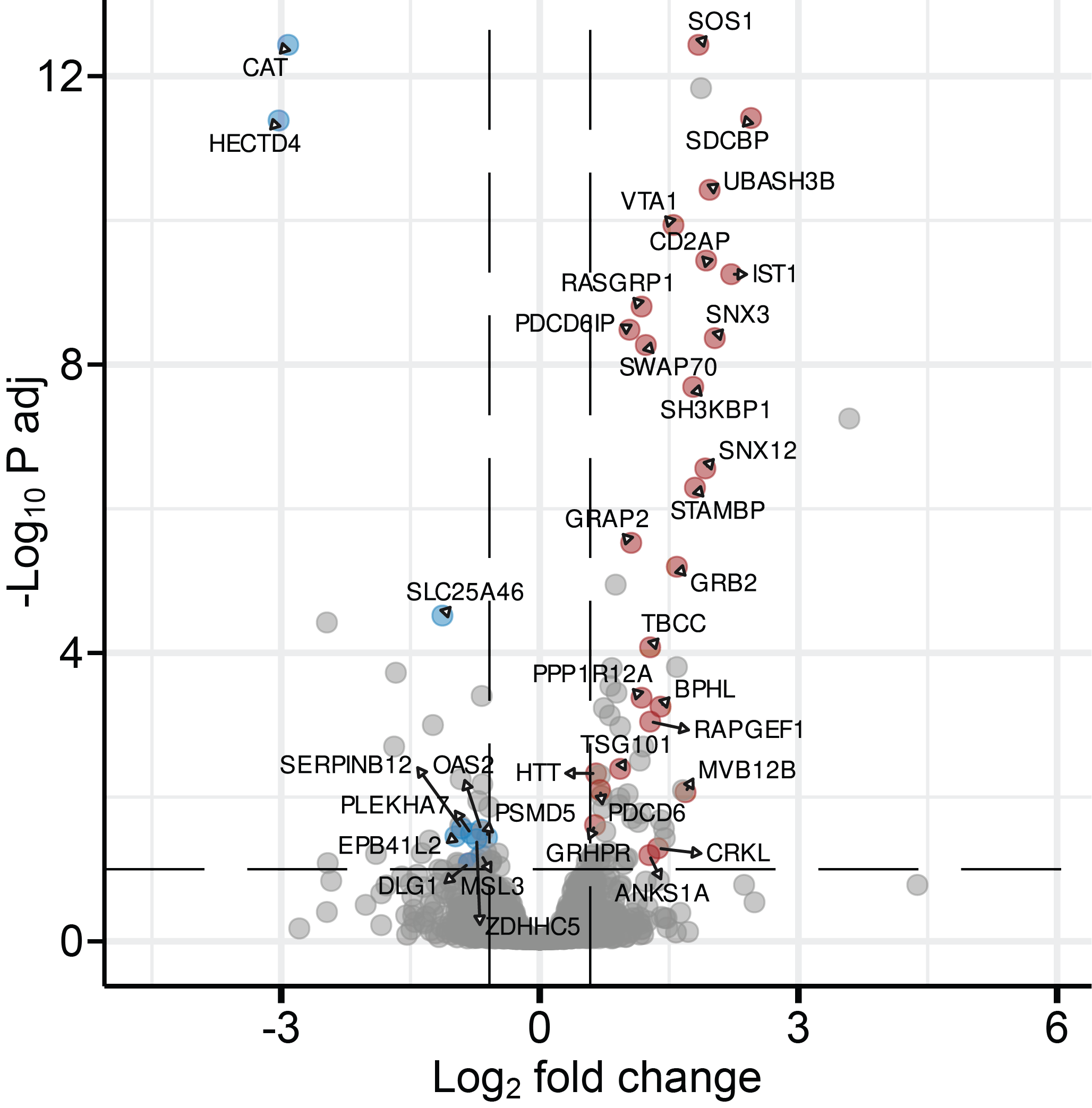
Volcano plot showing TIGIT interactome upon ligation vs non-ligation (Zammit et al, 2025).
Nanoscale Biology
Molecules within and on the surface of cells interact with each other within nanoscale dimensions. We use high-powered microscopes to reveal the ultrastructure of immune cell signalling. We also use super-resolution proximity proteomics to find interactions with high precision.
Nanoscopic resolution allows us to see fine details of molecular signalling, with greater depth and accuracy. It enables us to see processes that can’t be seen with normal microscopic resolution. These new insights will allow us to gain a deeper understanding of biological processes, driving the development of novel therapies.

Our Leica GSD nanoscope, for performing TIRF, STORM and 3D-STORM.
Publications
2025
Zammit, W.H, Le Maistre, L., Elliot, T.A.E., Cain, S.A.,Humphries, M.J.*, Davis, D.M.*, Worboys, J.D.*, Inhibitory TIGIT signalling is dependent on T cell receptor activation. BioRxiv, 2025.05.08.652881
Rey, C.*, Jones, K.L.*, Stacey, K.B., Evans, A.R., Worboys, J.D., Howell, G.J. and Davis, D.M., CD8α and CD70 mark human natural killer cell populations which differ in cytotoxicity. Frontiers in Immunology, 16, p.1526379.
2024
Worboys, J.D. and Davis, D.M., 2024. Do inhibitory receptors need to be proximal to stimulatory receptors to function?. Genes & Immunity, pp.1-3.
Shirgill, S., Nieves, D.J., Pike, J.A., Ahmed, M.A., Baragilly, M.H., Savoye, K., Worboys, J., Hazime, K., Garcia, A., Williamson, D.J., Rubin-Delanchy, P., et al. 2024. Nano-org, a functional resource for single-molecule localisation microscopy data. bioRxiv, pp.2024-08.
2023
Worboys, J.D., Vowell, K.N., Hare, R.K., Ambrose, A.R., Bertuzzi, M., Conner, M.A., Patel, F.P., Zammit, W.H., Gali-Moya, J., Hazime, K.S., Jones, K.L., et al. 2023. TIGIT can inhibit T cell activation via ligation-induced nanoclusters, independent of CD226 co-stimulation. Nature Communications, 14(1), p.5016.
Worboys, J.D., Palazón-Riquelme, P. and López-Castejón, G., 2023. Method to Measure Ubiquitination of NLRs. In NLR Proteins: Methods and Protocols (pp. 105-114). New York, NY: Springer US.
2022
Dechantsreiter, S.*, Ambrose, A.R.*, Worboys, J.D., Lim, J.M., Liu, S., Shah, R., Montero, M.A., Quinn, A.M., Hussell, T., Tannahill, G.M. and Davis, D.M., 2022. Heterogeneity in extracellular vesicle secretion by single human macrophages revealed by super‐resolution microscopy. Journal of Extracellular Vesicles, 11(4), p.e12215.
2021
Diaz-del-Olmo, I., Worboys, J., Martin-Sanchez, F., Gritsenko, A., Ambrose, A.R., Tannahill, G.M., Nichols, E.M., Lopez-Castejon, G. and Davis, D.M., 2021. Internalization of the membrane attack complex triggers NLRP3 inflammasome activation and IL-1β secretion in human macrophages. Frontiers in Immunology, 12, p.720655.
2020
Ambrose, A.R.*, Hazime, K.S.*, Worboys, J.D., Niembro-Vivanco, O. and Davis, D.M., 2020. Synaptic secretion from human natural killer cells is diverse and includes supramolecular attack particles. Proceedings of the National Academy of Sciences, 117(38), pp.23717-23720.
Smith, S.L., Kennedy, P.R., Stacey, K.B., Worboys, J.D., Yarwood, A., Seo, S., Solloa, E.H., Mistretta, B., Chatterjee, S.S., Gunaratne, P. and Allette, K., 2020. Diversity of peripheral blood human NK cells identified by single-cell RNA sequencing. Blood advances, 4(7), pp.1388-1406.
2014-2018
Palazón‐Riquelme, P.*, Worboys, J.D.*, Green, J., Valera, A., Martín‐Sánchez, F., Pellegrini, C., Brough, D. and López‐Castejón, G., 2018. USP7 and USP47 deubiquitinases regulate NLRP3 inflammasome activation. EMBO reports, 19(10), p.e44766.
Tape, C.J., Ling, S., Dimitriadi, M., McMahon, K.M., Worboys, J.D., Leong, H.S., Norrie, I.C., Miller, C.J., Poulogiannis, G., Lauffenburger, D.A. and Jørgensen, C., 2016. Oncogenic KRAS regulates tumor cell signaling via stromal reciprocation. Cell, 165(4), pp.910-920.
Locard-Paulet, M., Lim, L., Veluscek, G., McMahon, K., Sinclair, J., Van Weverwijk, A., Worboys, J.D., Yuan, Y., Isacke, C.M. and Jørgensen, C., 2016. Phosphoproteomic analysis of interacting tumor and endothelial cells identifies regulatory mechanisms of transendothelial migration. Science signaling, 9(414), pp.ra15-ra15.
Kümper, S., Mardakheh, F.K., McCarthy, A., Yeo, M., Stamp, G.W., Paul, A., Worboys, J., Sadok, A., Jørgensen, C., Guichard, S. and Marshall, C.J., 2016. Rho-associated kinase (ROCK) function is essential for cell cycle progression, senescence and tumorigenesis. elife, 5, p.e12203.
Tape, C.J., Worboys, J.D., Sinclair, J., Gourlay, R., Vogt, J., McMahon, K.M., Trost, M., Lauffenburger, D.A., Lamont, D.J. and Jørgensen, C., 2014. Reproducible automated phosphopeptide enrichment using magnetic TiO2 and Ti-IMAC. Analytical chemistry, 86(20), pp.10296-10302.
Worboys, J.D., Sinclair, J., Yuan, Y. and Jørgensen, C., 2014. Systematic evaluation of quantotypic peptides for targeted analysis of the human kinome. Nature methods, 11(10), pp.1041-1044.
Tape, C.J., Norrie, I.C., Worboys, J.D., Lim, L., Lauffenburger, D.A. and Jørgensen, C., 2014. Cell-specific labeling enzymes for analysis of cell–cell communication in continuous co-culture. Molecular & Cellular Proteomics, 13(7), pp.1866-1876.
Funding

